Projects
-
Expanding Galaxy Europe and Bioconda (2022–present).
We contribute state-of-the-art image analysis tools to the Galaxy Project and Conda ecosystem, to make recent image analysis methods accessible for everyone, and to facilitate further research.
-
Image Analysis for Visual Inspection of Electric Generators (2019–present).
We are concerned with the development of methods for large-scale image stitching of video data, acquired by small robots under low-light conditions to facilitate the inspection process of electric generators.
-
Globally Optimal Segmentation using Shape and Intensity Information (2016–present).
We develop new globally optimal model-based methods for segmentation of cell nuclei. Our latest method that is SuperDSM, leverages superadditivity and deformable shape models. The method was published in IEEE TPAMI and presented at the DAGM GCPR.
Affiliations
-
Biomedical Computer Vision Group (BMCV),
Heidelberg University,
- Research Fellow, PhD Student (2016–2022),
- Postdoctoral Researcher (2022–present).
-
Forschungszentrum Jülich GmbH,
- ELIXIR Germany (2023).
-
Heinrich-Heine-Universität Düsseldorf,
- Student of Computer Science (2013–2016).
- M.Sc. Project: Blind Deconvolution of Noisy Image Sequences
-
MediTEC, RWTH Aachen University,
- Apprentice (2010–2013),
- Student Assistant (2013–2015).
-
University of Applied Sciences Aachen,
- Student of Scientific Programming (2010–2013).
- B.Sc. Thesis: Virtual Coronary Angiography based on CT Data
Talks and Posters
- Invited talk. European Galaxy Days: Streamlining the User Experience for Reproducible Image Analysis in Galaxy. Freiburg, Germany, Oct 2025.
- Talk. Galaxy and Bioconductor Community Conference: Streamlining the User Experience for Reproducible Image Analysis in Galaxy. Cold Spring Harbor, New York, USA, Jun 2025.
- Talk. Galaxy Community Conference: Image Analysis in Galaxy: Recept Developments and Highlights. Brno, Czech Republic, Jun 2024.
- Poster. IEEE International Symposium on Biomedical Imaging: Robust Graph Pruning for Efficient Segmentation and Cluster Splitting of Cell Nuclei using Deformable Shape Models. Athens, Greece, May 2024.
- Invited talk. Next Generation Bioimage Analysis Workflows Hackathon: Workflows for Image Analysis using Galaxy. Irchel Campus, University of Zürich, Nov 2023.
- Poster. DAGM German Conference on Pattern Recognition: Superadditivity and Convex Optimization for Globally Optimal Cell Segmentation using Deformable Shape Models. Heidelberg, Germany, Sep 2023.
- Talk. International Conference on Control, Automation and Systems: Globally Optimal and Scalable Video Image Stitching for Robotic Inspection of Electric Generators. Jeju, Korea (online participation), Oct 2021.
- Talk. IEEE International Symposium on Biomedical Imaging: Segmentation of Cell Nuclei using Intensity-based Model Fitting and Sequential Convex Programming. Washington D.C., USA, Apr 2018.
Mentorship and Teaching
-
Series of Webinar Contributions on Image Analysis in Galaxy (2025) for GloBIAS Bioimage Analysis Seminar Series, EMBL EBI Course on Microscopy Data Analysis, and Euro-BioImaging Image Data Community Days 2025.
-
Organization of (2023) and Teaching within (2019, 2023) the training course “Microscopy Image Analysis”, de.NBI Project, HD-HuB.
-
Annual supervision of seminar projects (2022–present) on large-scale image analysis using Python for undergraduate students.
-
Supervision of student projects:
- J. Jegelka, “Interpretation of Machine Learning Models for Classification of Cell Nuclei and Imaging Artifacts” (2020–2021).
- S. Kopetschke, “Comparison of Different Image Stitching Software and Methods for Microscopy Images” (2019).
- R. Rappold, “Fluorescence Microscopy Image Data Search, Hyperparameter Optimization, and Comparison of Standard Methods for Cell Nuclei Segmentation” (2018).
-
Annual organization (2020–present) and supervision (2017–present) of programming classes using Java (2017–2019) and Python (2020–present) for undergraduate students.
Dr. Leonid Kostrykin is a postdoctral researcher in the Biomedical Computer Vision Group at Heidelberg University. His main interests are globally optimal methods for computer vision. As the ELIXIR Germany Officer of the Heidelberg Center for Human Bioinformatics (HD-HuB), he drives new developments of image analysis tools for Galaxy Europe and Bioconda.

Project on Image Analysis in Galaxy at the BioHackathon Europe 2025
Galaxy Project
November 2025. With the project entitled “Evolving FAIR image analysis in Galaxy for cross-domain and AI-ready applications” and lead by Dr. Kostrykin, it has been three years in a row that the community for image analysis in Galaxy succeeded with a project at the BioHackathon Europe:
- Enhancing the image analysis community in Galaxy (2023, online participation)
- Development of FAIR image analysis workflows and training in Galaxy (2024, nominated participant, Barcelona, Spain)
- Evolving FAIR image analysis in Galaxy for cross-domain and AI-ready applications (2025, project lead, Berlin, Germany)
The five-days hackathon is organized annualy by ELIXIR. Within the 2025 project, we have been able to align joint efforts from Euro-BioImaging, OSCARS FIESTA, and OSCARS BIO-CODES to further advance the ecosystem for image analysis in Galaxy.
Webinar Series on Image Analysis in Galaxy
Training Delivery
April 2025. This spring, we have delivered three webinars on image analysis in Galaxy. Our contributions for the GloBIAS Bioimage Analysis Seminar Series and the EMBL EBI Course on Microscopy Data Analysis were attended by respectively about 50 and 40 scientists from across the globe. The GloBIAS webinar was concluded by an extensive Q&A that is now available on the Image.SC Forums. Our third contriubtion was for the Euro-BioImaging Image Data Community Days 2025 and had about 90 attendees from across Europe.
Method and Device for Monitoring the State of a Component
Patent Granted
September 2024. Following its acceptance as an international patent, our patent application “Method and Device for Monitoring the State of a Component” has now also been registered as a US patent under the Publication Number US 2024/0296573 A1.
In 2023, our patent “Method and Device for Monitoring the State of a Component” was already granted and published under the Patent Cooperation Treaty (PCT) by the World Intellectual Property Organization (WIPO) under the International Publication Number WO 2023/025570 A1.
Both patents are based on our previous publication “Globally Optimal and Scalable Video Image Stitching for Robotic Inspection of Electric Generators”(Kostrykin et al. ICCAS 2021).
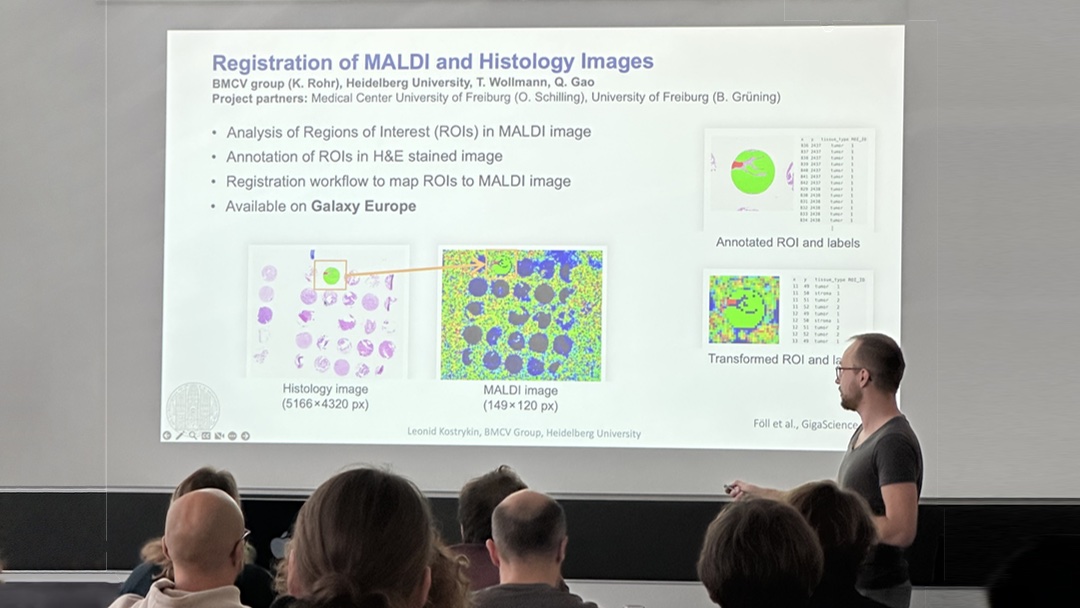
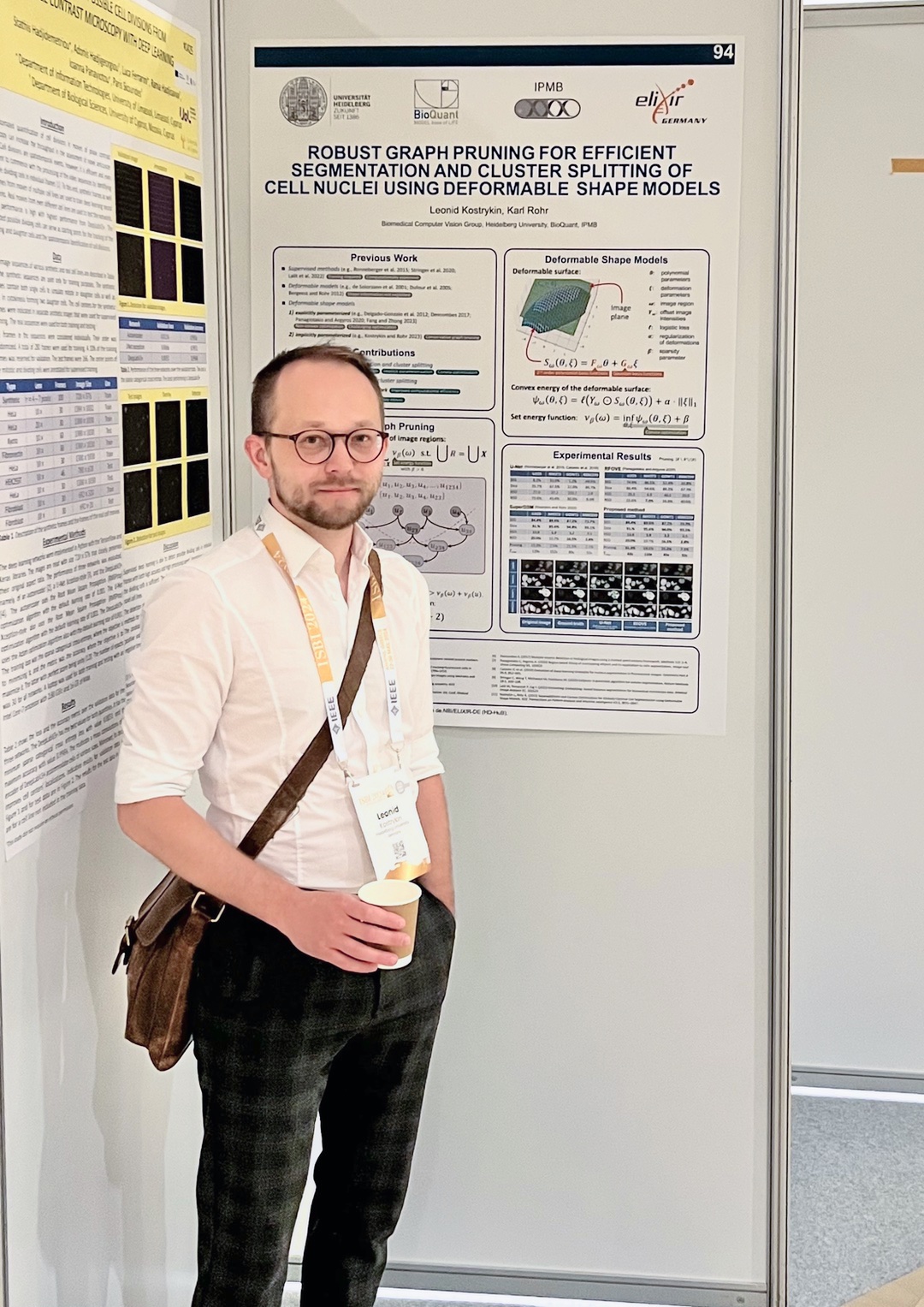
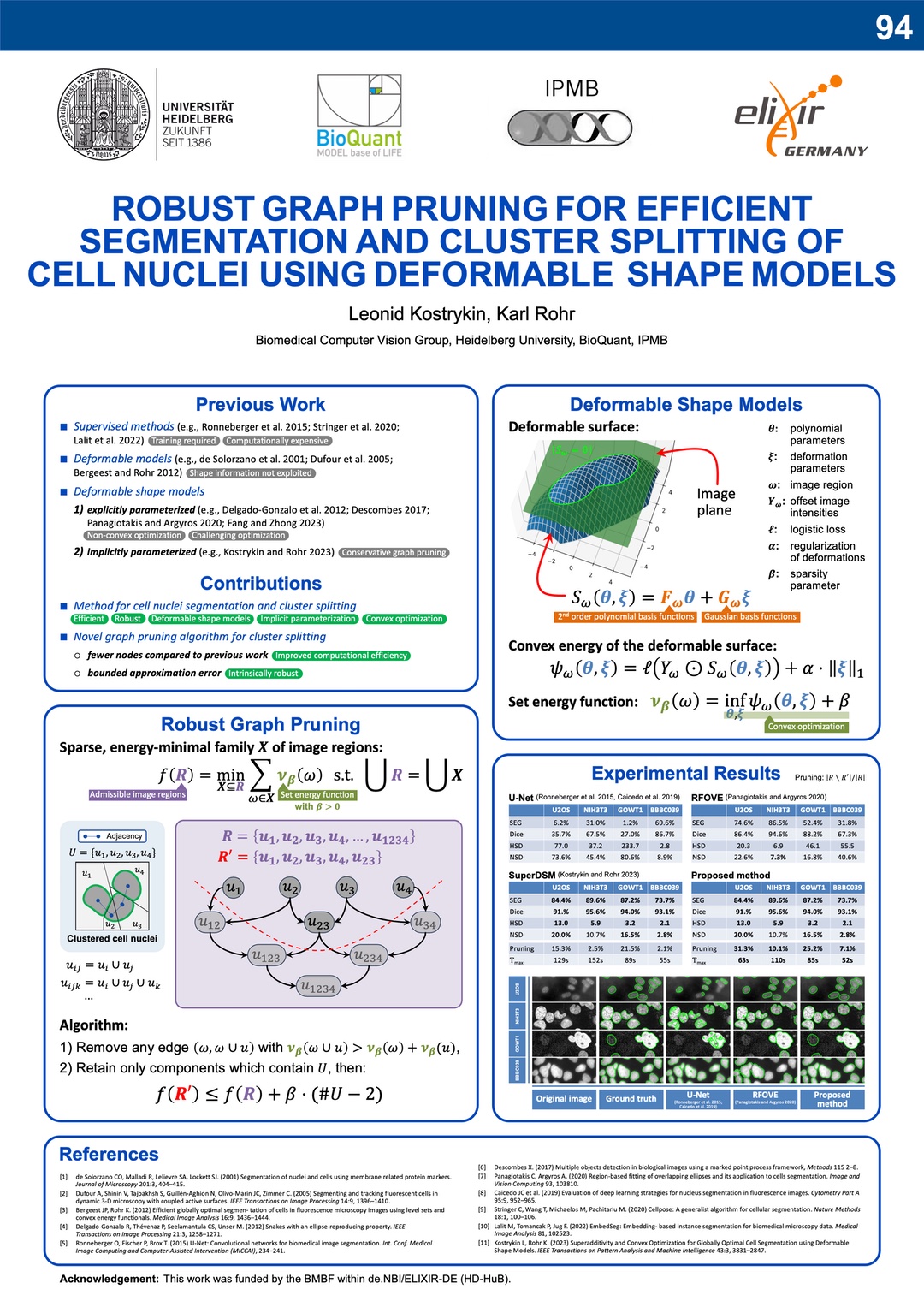
Robust Graph Pruning for Efficient Segmentation and Cluster Splitting of Cell Nuclei using Deformable Shape Models
L. Kostrykin and K. Rohr
Proc. IEEE International Symposium on Biomedical Imaging (ISBI 2024)
May 2024. Our conference paper “Robust Graph Pruning for Efficient Segmentation and Cluster Splitting of Cell Nuclei using Deformable Shape Models” has been presented at the IEEE ISBI 2024 in Athens, Greece. This contribution significantly improves the efficiency SuperDSM (our previously published method for cell nuclei segmentation, based on superadditivity and deformable shapre models).
The software is publically available on Galaxy Europe, Bioconda, and GitHub.
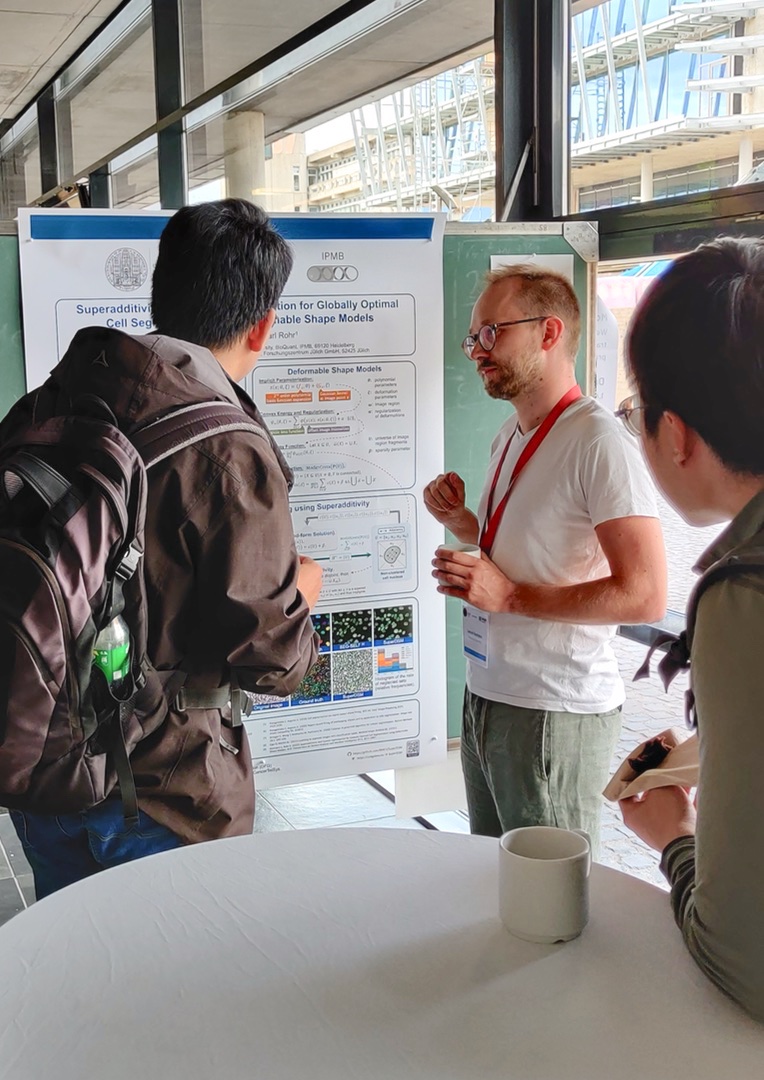
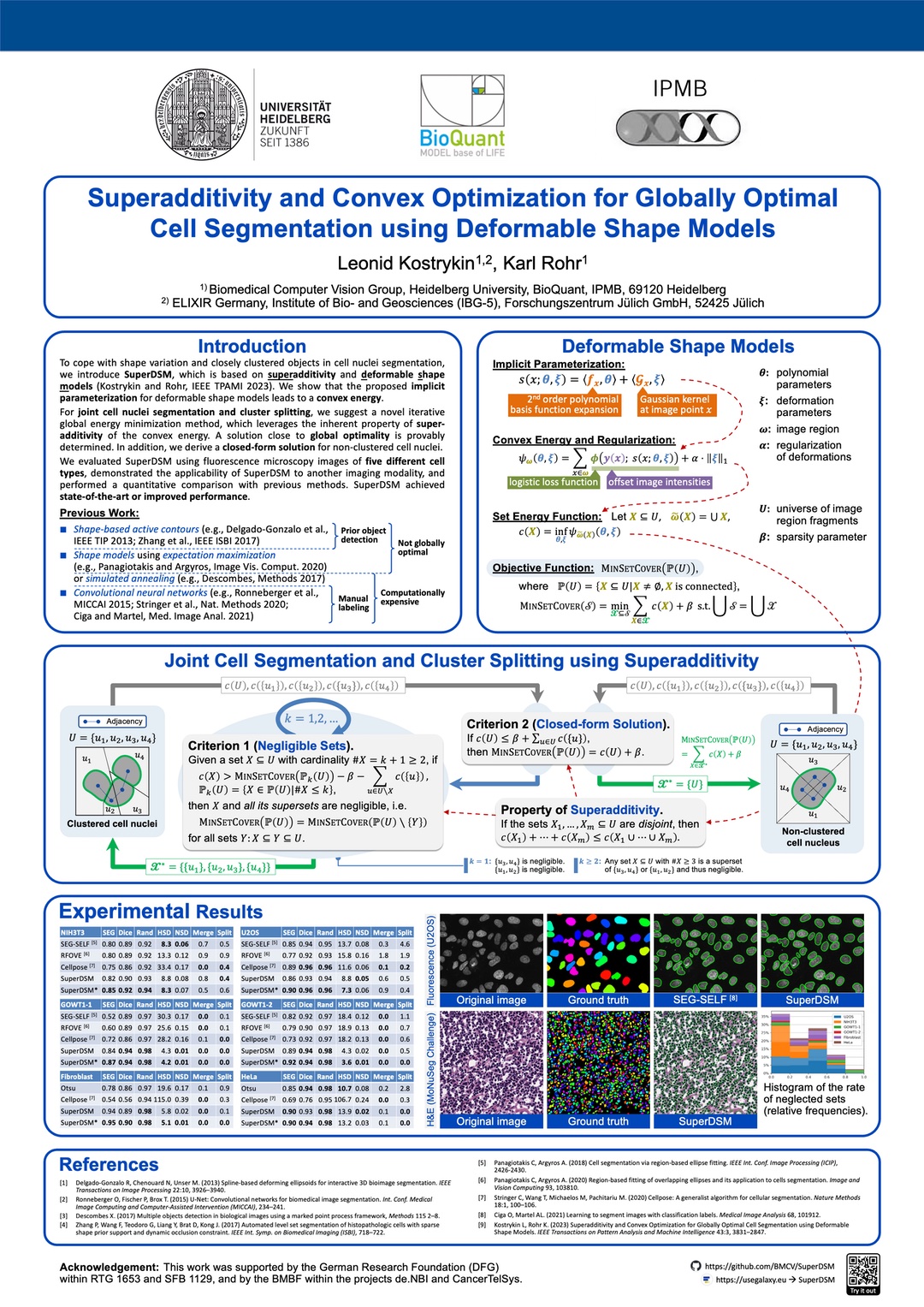
Superadditivity and Convex Optimization for Globally Optimal Cell Segmentation using Deformable Shape Models
L. Kostrykin and K. Rohr
IEEE Transactions on Pattern Analysis and Machine Intelligence
March 2023. Our article “Superadditivity and Convex Optimization for Globally Optimal Cell Segmentation Using Deformable Shape Models” has been published in IEEE TPAMI (vol. 45, no. 3). The article introduces SuperDSM, a new globally optimal method for segmentation of cell nuclei using superadditivity and deformable shape models. Some corrections can be found here.
SuperDSM is publically available on Galaxy Europe, Bioconda, and GitHub. The work has also been presented on the Nectar Track of the German Conference for Pattern Recognition (DAGM GCPR 2023) in Heidelberg, Germany.

Microscopy Image Analysis Course 2023
Training Delivery
August 2023. We have delivered a training course on Microscopy Image Analysis. In this course, we gave an introduction into the field of microscopy image analysis using automated software tools (ImageJ and Galaxy). The course was attended by 17 participants, including scientists from Germany and Italy.
Links: https://www.bioquant.uni-heidelberg.de/groups/rohr/micria-2023 https://www.denbi.de/training-archive-sorted-according-by-date/1574-microscopy-image-analysis-course-2023